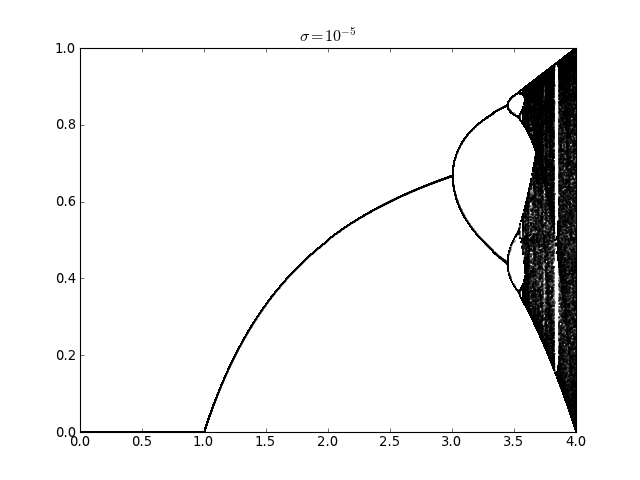
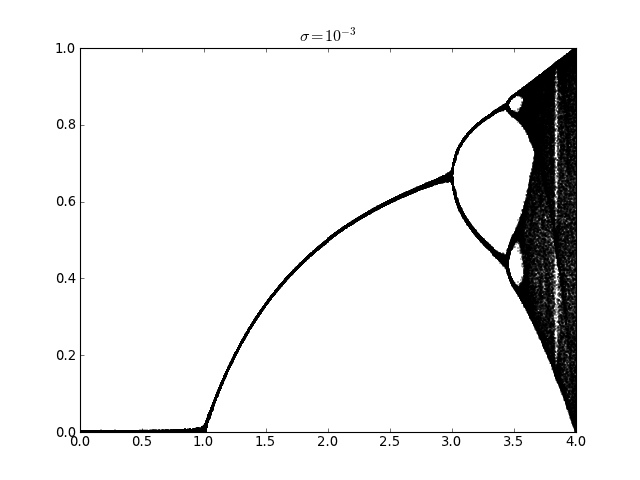
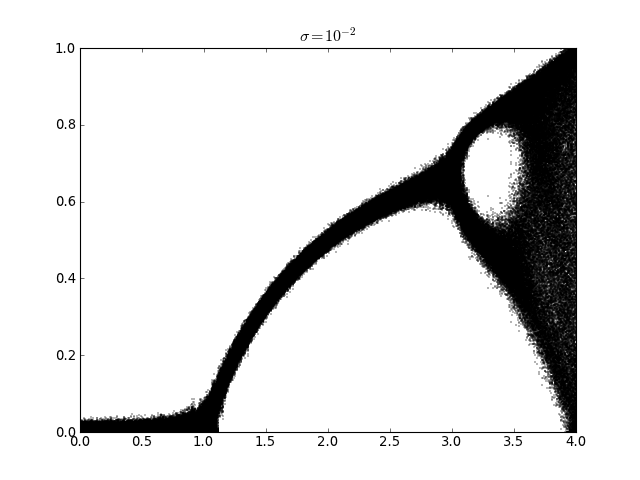
Logistic map with additive noise (usage of railgun.cmem())¶
You will need the file gslctypes_rng.py (see Using RailGun with GSL (GNU Scientific Library)).
Python code¶
logistic_map.py:
from railgun import SimObject, relpath, cmem
import numpy
from gslctypes_rng import gsl_rng
class LogisticMap(SimObject):
_clibname_ = 'liblogistic_map.so'
_clibdir_ = relpath('.', __file__)
_cmembers_ = [
'num_i',
'double xt[i]',
'double mu',
'double sigma',
cmem(gsl_rng, 'rng'),
]
_cfuncs_ = ["xt gene_seq()"]
def __init__(self, num_i, seed=None, **kwds):
SimObject.__init__(self, num_i=num_i, rng=gsl_rng(seed), **kwds)
def _cwrap_gene_seq(old_gene_seq):
def gene_seq(self, **kwds):
# use the previous "last state" as initial state, but setv
# may overwrite this by `xt_0=SOMEVAL`
self.xt[0] = self.xt[-1]
self.setv(**kwds)
return old_gene_seq(self)
return gene_seq
def bifurcation_diagram(self, mu_start, mu_stop, mu_num, **kwds):
mu_list = numpy.linspace(mu_start, mu_stop, mu_num)
self.gene_seq(mu=mu_list[0], **kwds) # through first steps away
xt_list = [self.gene_seq(mu=mu).copy() for mu in mu_list]
return (mu_list, xt_list)
def plot_bifurcation_diagram(self, mu_start, mu_stop, mu_num, **kwds):
import pylab
bd = self.bifurcation_diagram(mu_start, mu_stop, mu_num, **kwds)
ones = numpy.ones(self.num_i)
for (mu, x) in zip(*bd):
pylab.plot(ones * mu, x, 'ko', markersize=0.5)
pylab.ylim(0, 1)
bifurcation_diagram.py:
from logistic_map import LogisticMap
import pylab
lmap = LogisticMap(1000)
pylab.figure(1)
lmap.plot_bifurcation_diagram(0, 4, 500, sigma=1e-5)
pylab.title(r'$\sigma=10^{-5}$')
pylab.figure(2)
lmap.plot_bifurcation_diagram(0, 4, 500, sigma=1e-3)
pylab.title(r'$\sigma=10^{-3}$')
pylab.figure(3)
lmap.plot_bifurcation_diagram(0, 4, 500, sigma=1e-2)
pylab.title(r'$\sigma=10^{-2}$')
pylab.show()
C code¶
logistic_map.c:
#include <gsl/gsl_rng.h>
#include <gsl/gsl_randist.h>
typedef struct logisticmap_{
int num_i;
double *xt;
double mu;
double sigma;
gsl_rng *rng;
} LogisticMap;
int LogisticMap_gene_seq(LogisticMap *self)
{
int i;
double eta;
for (i = 1; i < self->num_i; ++i){
eta = gsl_ran_gaussian_ziggurat(self->rng, self->sigma);
self->xt[i] = self->mu * self->xt[i-1] * (1 - self->xt[i-1]) + eta;
if (self->xt[i] < 0){
self->xt[i] = 0;
}
}
return 0;
}